July 2012
By Colleen Daniels and Coco Jervis
Introduction
Accurate tuberculosis (TB) diagnosis has the potential to be the cornerstone of all global TB control efforts. However, the poor accuracy of the most commonly used test, sputum smear microscopy, and the weeks-long time to results of the previously prevalent gold standard for TB diagnosis of culture growth, have combined to make accurate diagnosis the Achilles’s heel of efforts to eradicate TB. The World Health Organization (WHO) estimates that one-third, or almost 3 million, of the world’s 8.8 million TB cases are never detected or reported let alone diagnosed, treated, and cured properly.[1] The most commonly used TB tests involve a trade-off between insensitive assays such as microscopy, which detects only 50% of all TB cases, and fewer in children or people with HIV, and accurate ones (such as culture) that require weeks of in vitro growth before generating results, meaning that people with possible TB must come back to the lab weeks later to begin appropriate treatment. In some places such as the private sector in India, people are often given ineffective and expensive TB antibody tests that combine uselessness with profits to the lab owners, with no benefit to the patient. These barriers ensure that many patients are neither diagnosed nor treated properly.
The past decade has seen both the rollout to developing countries of existing technologies such as rapid liquid culture and the development of new molecular (DNA-detecting) technologies such as the Hain Lifescience (Germany) GenoType MTBDRsl and the Cepheid (United States) GeneXpert that detect both the presence of TB and common resistance mutations to isoniazid and rifampicin. This progress, however, must be tempered by the recognition that we still do not have a low-cost, laboratory-free point-of-care (POC) test for TB.[2] We have seen how POC tests for “HIV and malaria have completely transformed the management of these diseases,”[3] and we now wait impatiently for one to change the management of TB.
The need for simpler methods to accurately diagnose TB in everyone including children and people with immune suppression such as HIV cannot be overstated—the faster a patient is screened, the quicker treatment can be initiated, ultimately reducing morbidity and reducing the spread of TB.[4]
Increased funding for TB research and development is still desperately needed in order to maximize the breadth and depth of innovative technologies currently being considered in the TB diagnostic pipeline. Funding for TB diagnostics research was a minimal US$44.6 million in 2010,[5] compared to the US$340 million the Global Plan to Stop TB indicates is needed annually.[6]
Factors essential for an improved TB diagnostic test
The utilityof a test is defined by the following factors linked to test accuracy and the place within the health system where the test is likely to be used.
Sensitivity:The ability of the test to accurately identify people with the disease. Low sensitivity of a test will cause people who have the disease to not be identified, not get appropriate treatment, suffer due to disease progression, and transmit the disease to others.
Specificity:The ability of the test to accurately identify people who do not have the disease. Low specificity means that more people who do not have the disease will wrongly be identified as having it, leading to inappropriate treatment.
Impact of test results on clinical decisions and patient outcomes: Sensitivity and specificity are surrogates for a test’s ability to improve treatment outcomes. Even a highly sensitive and specific test may not result in improved treatment decisions or reduce morbidity and mortality if it takes too long to provide results, thus failing to allow prompt initiation of proper treatment.[7]
Diagnostic algorithm: An algorithm is a recommended sequence in which procedures such as symptom screens can be combined with tests in a diagnostic pathway. The most efficient programs strive to integrate procedures and tests to ensure the most rapid, accurate, and rational diagnosis and treatment for all patients.
Health posts: These are the most decentralized locations of the health system, serving 60% of TB patients. They often lack access to electricity, water, or trained laboratory staff, and do not support diagnostic or biosafety equipment.
Peripheral laboratories or health centers: These include district hospitals and laboratories and serve 25% of people in need of TB services. They have trained staff and the capacity to conduct sputum smear microscopy, but only inconsistent electricity and minimal biosafety capacity.
Reference laboratories with sophisticated test procedures are accessible to only 15% of those in need of TB services.They have highly skilled staff, reliable electricity and water supply, can ensure biosafety, and can conduct culture and nucleic acid amplification tests (NAATs).[8]
Source: 2011 Pipeline Report.[9]
As a result of these continuing discussions, this year’s pipeline is structured by the diagnostic test technology: culture, molecular, and non-molecular.
The TB Diagnostics Pipeline
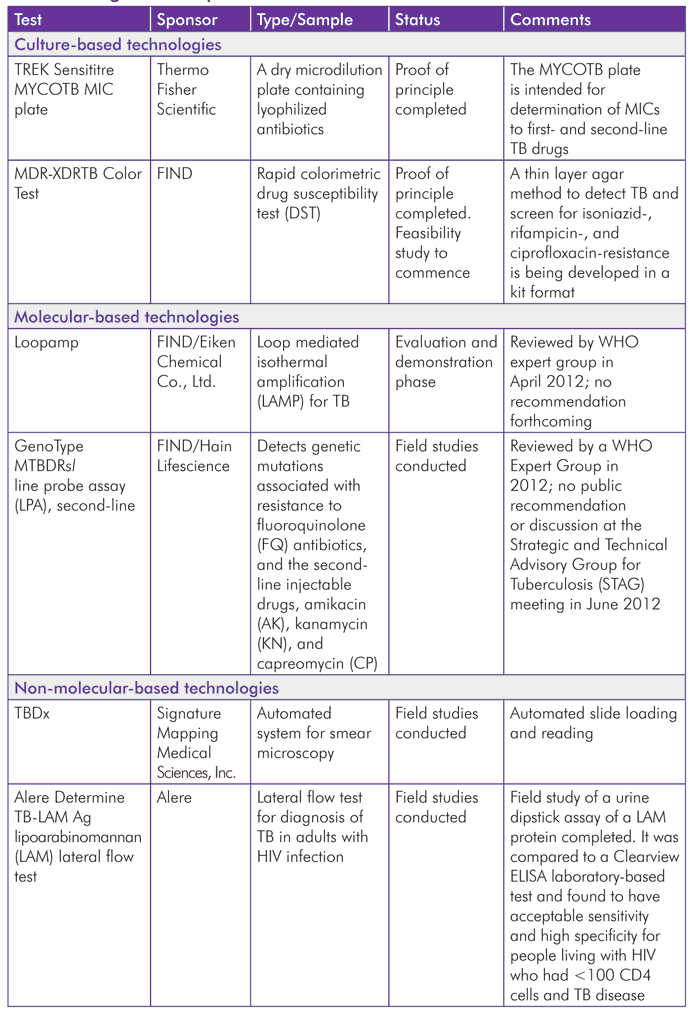
Culture Technologies
The most widely used technology to diagnose TB is acid-fast bacilli (AFB) sputum smear microscopy. It is rapid and inexpensive, but has low sensitivity, particularly in people living with HIV (PLHIV). It also does not provide drug susceptibility information, and performance of the test depends on the operator.[10],[11] Liquid culture media are more sensitive and faster than traditional techniques, which use egg-based solid media. However, culture using thin layer synthetic agar (which uses a solid medium and is based on the microscopic detection of early mycobacterial growth)[12] does improve performance of solid culture media.[13] Liquid culture systems reduce delays in getting results, and for DST the result can be obtained in less than 10 days compared to 28–42 days for solid media.[14] Liquid systems have more issues with contamination and require a strong network of quality-assured microscopy in countries. The WHO recommends the use of TB liquid culture and DST in low-income settings and implementation of these systems as part of national laboratory strengthening plans.[15]
There are two culture-based technologies in the pipeline: the TREK Sensititre MYCOB MIC plate and the solid culture MDR/XDR Color Test.
TREK Sensititre MYCOTB MIC Plate
The TREK Sensititre MYCOTB MIC (Trek Diagnostic Systems, Thermo Fisher Scientific, United States) plate is intended for determination of drug resistance to first- and second-line TB drugs. A multisite study was conducted by the TB Clinical Diagnostics Research Consortium (CDRC) to evaluate the diagnostic accuracy of the MYCOTB plate compared to the reference agar proportion method (APM). Two reference TB laboratories, one in South Korea and one in Uganda, conducted the study.
A presentation by Dr. Susan Dorman[16] at the Keystone Symposium on Drug Resistance and Persistence held in Uganda in May 2012, compared APM with TREK in archived Mycobacterium tuberculosis (MTB) clinical strains from South Korea and Uganda with previously characterized DST patterns were used. A “total of 228 MTB isolates were selected, among which 69 (30%) had previously been characterized as MDR-TB and 52 (23%) had been characterized as XDR-TB. The MYCOTB plate showed ≥95% concordance with APM for all tested drugs except ethionamide 5.0 ug/mL (94.0%), rifabutin (92.4%), moxifloxacin 0.5 ug/mL (87.3%), and moxifloxacin 2.0 ug/mL (81.0%) in interim results. MYCOTB plate interpretation by two independent readers showed ≥96% agreement for all tested drugs.”[17]
Solid Culture MDR-XDRTB Color Test
A thin layer agar method[18] (TLA) to detect TB and screen for isoniazid-, rifampicin-, and ciprofloxacin-resistance is being developed in a kit format by the Foundation for Innovative New Diagnostics(FIND) and the London School of Hygiene & Tropical Medicine (LSHTM). The color test makes it possible to perform sputum smear and culture plate inoculation and incubation safely in basic regional laboratories that currently provide only sputum microscopy, with biosafety requirements that are similar to those for smear microscopy. Two drops of a sample are dropped onto selective thin layer agar for incubation in room air, resulting in MDR-TB (multidrug-resistant TB) testing and XDR-TB (extensively drug-resistant TB) screening.[19]
FIND and LSHTM established proof-of-concept for this approach. A prospective feasibility study using sputum is now ready for enrolment, and will be conducted in a regional laboratory in Brasov, Romania, and in a referral laboratory in Lima, Peru. The study aims to evaluate the operational feasibility of using sputum from TB suspects, and will compare performance of the color test for TB- and resistance-detection against the automated mycobacteria growth indicator tube (MGIT) system and the Löwenstein-Jensen (LJ) culture systems.[20]
Molecular-Based Technologies
Molecular-based detection of TB includes the use of nucleic acid amplification tests (NAATs), which are specific and reliable. Test results are normally available within 24 to 48 hours—and sometimes within two hours—of receipt of sample, compared to culture, which takes weeks.[21] Sensitivity, however, is variable, especially in smear negative and extrapulmonary samples.
Line probe assay technology used to detect MDR-TB involves extracting DNA from MTB isolates or directly from clinical specimens. Polymerase chain reaction (PCR) amplification of the resistance-determining region of the gene is performed. The readout shows whether the sample contains MTBand whether defined resistance sequences are present.[22]
Manual Isothermal NAAT for TB (LAMP)
FIND (Switzerland) and Eiken Chemical Co., Ltd. (Japan) are developing a loop-mediated isothermal amplification (LAMP) technology for TB detection in sputum using a relatively convenient kit format (Loopamp) with a simple visual readout. Evaluation and demonstration study phases were completed in 2011 at three sites in India, Uganda, and Peru. The Indian results were discordant with the other two and with test specifications. As a result, the phase I demonstration study will be redone to prove/disprove the hypothesis that the cause of the different results was due to a need for greater training. Eiken and FIND will redo the evaluation study at all sites (South Africa, Vietnam, Brazil, and Peru) to include the updated training module.[23] The new device includes a specially designed sputum-transfer device to ensure that appropriate volumes of sputum will be supplied with the finished kit. While a WHO expert group reviewed the data on LAMP and Loopamp in April 2012, no public recommendation or discussion occurred at STAG in June.
Molecular Detection of Drug Resistance
GenoType MTBDRsl
Based on the already-approved GenoType MTBDRplus, which detects common isoniazid- and rifampicin-resistance-associated mutations, the Hain PCR-based assay GenoType MTBDRsl allows for the simultaneous detection of TB organismand its resistance to fluoroquinolones, aminoglycosides/cyclic peptides, and ethambutol.[24] It is used to detect XDR-TB. Several studies have assessed this test.
At four sites in Eastern Europe, Ignatyeva et al.[25] evaluated the performance of the Genotype MTBDRsl assay compared to that of phenotypic drug susceptibility testing (Becton Dickinson’s BACTEC MGIT 960 system). Sensitivity for the detection of resistance to fluoroquinolones, ethambutol, amikacin, and capreomycin varied between 77.3% and 92.3% and was much lower for kanamycin at 42.7%. The sensitivity for the detection of XDR-TB was 22.6%, and test specificity was over 82% for all drugs.[26]
In South Africa, Said et al.[27] found that the sensitivity and specificity of the GenoType MTBDRsl assay were, respectively, “70.3% and 97.7% for ofloxacin, 25.0% and 98.7% for kanamycin, 21.2% and 98.7% for capreomycin and 56.3% and 56.0% for ethambutol.”[28] The assay performed well for ofloxacin, was less sensitive for kanamycin- and capreomycin-resistance, and had low sensitivity and specificity for ethambutol-resistance.
The study conducted in Eastern Europe concluded that the sensitivity for the detection of kanamycin-resistance needs improvement, and the South African study recommended that the GenoTypeMTBDRsl assay include additional genes to achieve better sensitivity for all the drugs tested.
Last year, a WHO Expert Group (EG) considered data from this and unpublished studies and determined that the available data supported a recommendation for use of the assay testing culture isolates, but that it could not endorse the use of the assay for direct testing on sputum specimens because there were too few data on direct testing available. As a result, FIND and Hain Lifescience GmbH (developer and manufacturer) implemented further studies of direct testing. These data on the GenoType MTBDRsl test were presented to a WHO EG in April 2012. Neither a recommendation nor a public explanation of the current WHO evaluation of this test were available publicly as this report went to press.[29],[30]
Non-Molecular Diagnostic Technologies
Biomarkers are biologicfeatures that can identify and/or be used to monitor a physiological process or disease in the host. Nahid et al. reported that several new approaches to discovery of TB diagnostics biomarkers are now being researched with a focus on pathogen-specific or host-based markers. Researchers have screened urine, serum, saliva, and breath in the search for markers that can be evaluated via a variety of platforms including genomic, proteomic, metabolomic, lipidomic, and glycomic.[31]
TBDx
TBDx (Signature Mapping Medical Sciences, Inc., a subsidiary of Applied Visual Sciences, Inc., United States) is an automated system for smear microscopy that automatically loads and reads slides. It autofocuses and digitally captures images and uses computerized algorithms to count AFBs and classify slides as positive or negative.[32]
At the 2012Conference on Retroviruses and Opportunistic Infections (CROI), Dr. Gavin Churchyard presented results of a study, conducted by the Aurum Institute (South Africa) and Guardian Technologies International (United States), using culture as the control.[33] It concluded that the sensitivity of TBDx is very good, but specificity is too low, when used as a fully automated system. Test results by a microscopist showed sensitivity of 52.8% and specificity of 98.6% compared to TBDx, which had sensitivity of 75.8% and specificity of 43.5%. However, Dr. Churchyard also showed that TBDx tends to overread, which leads to high false-positivity. He concluded that it is a promising technology that merits further optimization and evaluation.
Alere Determine TB-LAM Ag
Alere Determine TB-LAM Ag (Alere, United States) is a lateral flow test for detection of urinary lipoarabinomannan (LAM).[34] The LAM protein is shed from TB bacteriain people with TB disease.
Lawn et al. in South Africa assessed the diagnostic accuracy of the Alere Determine TB-LAM Ag for screening for HIV-associated pulmonary tuberculosis before antiretroviral therapy (ART). Their study found that it had acceptable sensitivity and very high specificity for people with TB disease who also had CD4 counts <100. These results did not differ statistically from the sensitivities obtained by testing a single sputum sample with the Xpert MTB/RIF assay.[35]
Peter et al.[36] found that LAM combined with smear microscopy was able to rule in TB in 71% of MTB culture-positive patients. This indicates that the LAM strip test may be a potentially useful rapid rule-in test for TB in hospitalized patients with advanced immunosuppression.
Dr. Susan Dormanpresented a study at CROI 2012[37] which found that the Alere Determine TB-LAM Ag, which detected TB intwo-thirds of TB patients with CD4 counts <100, may be a clinically useful adjunct to conventional TB diagnostic testing in patients with very low CD4 counts.[38]
For the first time, there might be a point-of-care TB test that works well in people with very advanced AIDS—a group that is difficult to diagnose with sputum smear microscopy, has high mortality from TB, and stands to benefit most from prompt initiation of ART.[39]
Volatile Organic Compound (VOC) Analysis
Volatile organic compounds (VOCs) in breath provide biomarkers of TB because MTB manufactures VOC metabolites detectable in the breath of infected patients.[40]
Gas Chromatography-Mass Spectrometry(GC-MS) Breath Analysis
Kolk et al.[41] investigated the potential of breath analysis by gas chromatography-mass spectrometry (GC-MS) to discriminate between samples collected prospectively from patients with suspected TB. With an accuracy of 79%, the results are similar to those of Phillips et al.[42] and Kolk.[43] GC-MS breath analysis can differentiate between TB and non-TB breath samples even among patients with a negative ZN sputum smear but a positive culture for MTB. The study concluded that more research into breath analysis is now needed.
BCA 5.0
Phillips et al.[44] evaluated the BCA 5.0 (Menssana Research Inc., United States) in the Philippines, the United Kingdom, and India. The study evaluated breath VOC biomarkers in people with active pulmonary TB. Breath samples are collected and analyzed by gas chromatography for the detection of VOCs. The investigators reported detection of active pulmonary TB with 80% accuracy, 71.2% sensitivity, and 72% specificity. However, 87% of positives were false, which indicates that further refinement of this approach will be necessary before any field use can occur.
Other Diagnostics in the Pipeline
Several other diagnostics may be in the pipeline, but have no published studies. The TrueNAT MTB test by Bigtec labs and Molbio Diagnostics (India) is said to detect TB DNA in sputum within one hour by processing sputum on a semiautomated, battery-operated, portable device.[45] The TrueNAT MTB test and devices are currently under clinical evaluation in two other locations in India and South Africa, and more studies are planned in 2012.
Genedrive is a point-of-need genotyping and sequence-analysis device being made by Epistem (United Kingdom). Epistem’s product information states that Genedrive has been designed as a handheld device that analyzesnucleic acids and proteins from fresh or stored biospecimens in clinical settings. Human TB validation studies are under way Spain, India, and South Africa. Epistem expects to be awarded CE/IVD accreditation (European Union accreditation for medical devices) in 2012.[46]
B-SMART (Sequella, Inc., United States) is a test designed to rapidly detect the presence of TB bacteriawith resistance to the four front line anti-TB drugs: rifampicin, isoniazid, streptomycin, and ethambutol[47] directly from sputum.[48] Sequella states that the B-SMARTprototype assay detects TB bacteriawith at least the sensitivity of the sputum smear (<1,000 cells). The developers hope that optimization will significantly increase sensitivity (<50 cells) in order to assess TB drug resistance in smear-negative, culture-positive clinical samples. A study is ongoing.
A project conducted by FIND (Switzerland) and MBio Diagnostics, Inc. (United States) designed to determine a set of serodiagnostic TB antigens for diagnosis of active disease, which will eventually result in a POC assay format, has progressed to the development of a platform POC test that will enter field evaluation in 2012. FIND and MBio are planning a clinical study at two field sites in developing countries.[49]
While many of the emerging technologies look promising, it remains to be seen if they actually live up to their potential in rigorous clinical and demonstration studies in routine programmatic settings. A recent study by Denkinger et al.[50] showed that optimism bias is a concern with package inserts of TB diagnostics (i.e., industry claims); they compared test accuracy for TB diagnostics reported in 19 package inserts against estimates in published meta-analyses, and found that package inserts generally report overoptimistic accuracy estimates. However, package inserts of most tests approved by the FDA or endorsed by the WHO provide more realistic estimates that agree with meta-analyses.
Global and National Diagnostics Policy Development
Over the past decade, at least 20 new diagnostic test platforms have been discovered, developed, and evaluated.[51]
Many countries will not adopt a test unless it is first recommended by the WHO, which endorses new tuberculosis diagnostics by using the Grading of Recommendations Assessment, Development and Evaluation (GRADE) process. This process is mostly based on test accuracy, with limited cost and feasibility data. The WHO’s 2010 Handbook for Guideline Development states that a “recommendation is then made which is strong or conditional/optional/weak (for or against an intervention).” The Handbook also states that the GRADE process takes into “account the benefits and downsides, values and preferences, impact, and resource use. This is balanced with the quality of evidence(high, moderate, low, very low), the methodological quality of evidence, any likelihood of bias and by outcome and across outcomes.”[52]
A new approach to developing policies is now being discussed, and Cobelens et al. have called for a revision of this system to speed adoption of new diagnostics for tuberculosis.[53] The proposed policy process is now envisioned as having two steps: an initial technical recommendation, followed by a programmatic recommendation. Cobelens et al. suggest that the technical recommendation would still follow the current GRADE process and be based on test accuracy with limited costing and feasibility data, however, programmatic recommendation would include patient-important outcomes, cost-effectiveness when implemented under routine conditions, and other factors critical to successful scale-up at the country level. The evidence for both steps should be systematically collected, but each requires different study designs.[54] The new value chain separates activities at the global level from activities that need to occur at the country level. It also involves multiple feedback loops whereby evidence at each level can inform subsequent decisions to scale up new technologies (or not), and decisions to modify or revise existing policies based on epidemiological impact (or lack thereof) at the country level.
Discussions around when to scale up new diagnostics and how to include them in diagnostic algorithms have increased recently, particularly in light of the rollout of GeneXpert.[55],[56] These discussions will ensure that appropriate polices that enable countries to implement rapidly are developed. However, we strongly recommend that the policy development process not allow a country to delay implementation until it has conducted its own field trials for every new test.
Xpert MTB/RIF Progress
A number of countries have been rolling out the Xpert MTB/RIF test following its endorsement by the WHO and the U.S. Federal Tuberculosis Task Force in 2010. GeneXpert is an automated diagnostic molecular testing system, which simultaneously detects TB and rifampicin drug resistance (using Xpert MTB/RIF cartridges) in less than two hours.[57] Widespread implementation of Xpert RIF/MTB began in 2011. According to the WHO and FIND, by March 2012, a total of 611 GeneXpert instruments (comprising 2,979 modules) and 863,790 Xpert MTB/RIF test cartridges had been procured in 61 countries under concessional pricing (WHO GeneXpert Update 2012).[58]
South Africa procured over half of the Xpert MTB/RIF cartridges (478,980); the rest were acquired by other countries as follows: Kenya (34,310); India (25,640); Pakistan (22,440); Zimbabwe (21,570); Tanzania (20,370); Nigeria (18,160); the Philippines (17,440); and Brazil (16,730).[59] At an April 2012 meeting hosted by the Stop TB Partnership Global Laboratory Initiative and the WHO in Annecy, France, Xpert MTB/RIF early implementers noted that the need to reduce the price of the Xpert MTB/RIF test cartridge from US$16.86to under US$10.00 is a priority, as participants saw cost as a major obstacle to an accelerated and sustainable rollout of the technology in low- and middle-income settings. In order to accelerate implementation, private-sector purchasers in many high-burden countries such as India and South Africa also need to be able to access the concessional prices offered to national TB programs, participants at the meeting said.[60]
Chang et al. reported on a meta-analysis to evaluate the rapid and effective diagnosis of tuberculosis and rifampicin resistance with Xpert MTB/RIF. The meta-analysis included “18 studies covering 10,224 specimens” and found that the accuracy of Xpert MTB/RIF in detecting pulmonary TB and RIF resistance was a “pooled sensitivity of 90.4%, pooled specificity of 98.4%; and for RIF, the pooled sensitivity was 94.1% and pooled specificity, 97%.” Xpert performance in detecting extrapulmonary TB was found to be pooled sensitivity of 80% and pooled specificity of 86%.[61]
Xpert MTB/RIF rollout: The South African experience
South Africa has procured over half of all GeneXpert machines and Xpert MTB/RIF cartridges to date. A pilot phase was initiated in National Health Laboratory Services (NHLS) microscopy centers in high-focus TB areas. A progress report on the implementation by the NHLS shows that at least one instrument was placed per province preferentially in districts that had a high burden of TB. The report stated that “twenty-five microscopy centers were selected and a total of 30 instruments placed.”[62]
According to the NHLS progress report, as of March 27, 2012, a total of 311,117specimens had been processed. The total percentage of TB detected in this cohort was between 16% and 17%; the national average was 16.74% (52,068 positive tests). Average rifampicin resistance-detection rates have remained around 7% since the start of the project.[63] A cluster-randomized pragmatic trial within the NHLS rollout of Xpert MTB/RIF will also be conducted in South Africa. It aims to evaluate the impact and cost-effectiveness of routine rollout of Xpert MTB/RIF. The study will also look at the impact on patient and program outcomes, and transmission at a population level. Patient outcomes will be measured for TB suspects and TB patients, and will include six-month mortality amongst TB suspects as the primary outcome. Upon completion of the study, researchers will also be able to determine whether the introduction of Xpert MTB/RIF alters provider behavior with respect to investigating TB suspects, and to estimate costs from the patients’ perspective.
Comprehensive economic costs (including costs to the health system) are also being measured, together with the parameters required for the modeling of population impact.For example, a study by Andrews et al.[64] based on modeling shows that Xpert MTB/RIF is cost-effective when using the diagnostic to screen individuals initiating ART. The study results indicate that when compared to no screening, life expectancy in patients with TB diseaseincreased by 1.6 months using smear in symptomatic patients, and by 6.6 months with two Xpert samples in all patients.[65]
Source: Regarding GeneXpert MTB/RIF progress report.[66]
Pediatric TB Diagnostic Update
Accurate diagnosis of TB in children poses multifaceted challenges that current diagnostic tools inadequately address. The childhood TB diagnostic research pipeline has been hampered by technical and clinical difficulties that have only recently been resolved. Chief among these challenges are the difficulty of obtaining sputum specimens from children, the slow growth of the organism in culture, and the diverse and relatively nonspecific clinical presentation of TB in children. Alternative samples for diagnostic testing in children through stool, urine, and saliva are now being explored, but nothing substantial has been advanced. Until recently, there was little consensus in the childhood TB research community on case definitions, as diagnostic classifications and reference standards vary dramatically among researchers.
Recently, the National Institutes of Health (NIH) set out to establish a new reference standard for the diagnosis of TB in children.[67] Leading childhood TB researchers and clinicians agreed on research reference standards and clinical case definitions for intrathoracic TB diagnosis in children. The experts also agreed to try to harmonize methodological approaches for evaluating new diagnosis tools in children.[68]
The consensus case definitions agreed to by the expert meeting are outlined below.
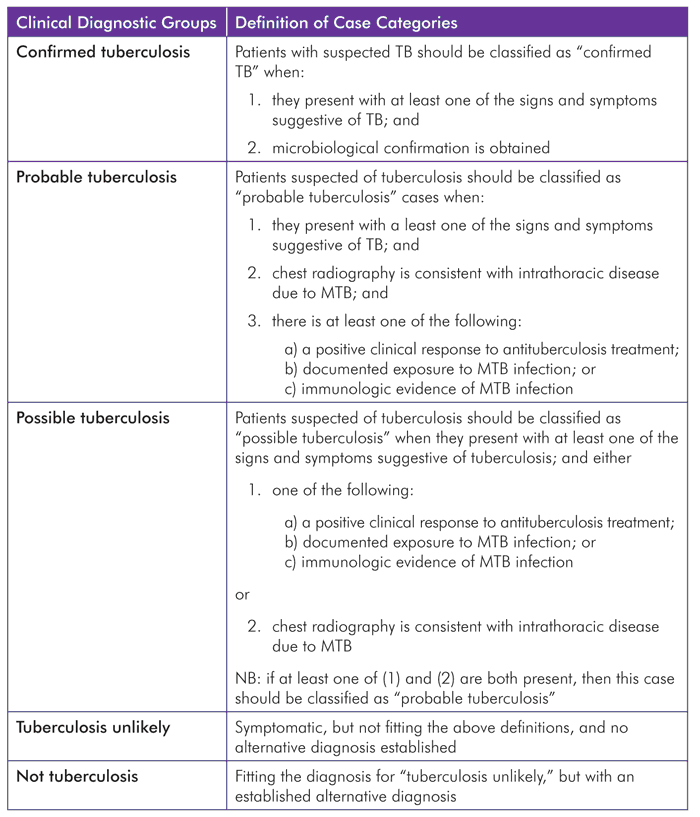
Source: Evaluation of tuberculosis diagnostics in children.[69]
Now that a pathway for classifying children with TB for diagnostic research purposes has been established, we can hope for better research and more data on new TB diagnostic tests for children in the near future. In a first-of-its-kind prospective study of 452 children under the age of 15 in Cape Town, South Africa, using Xpert MTB/RIF, accuracy exceeding that of microscopy was demonstrated in detecting TB in children.[70]
The study compared Xpert with culture using repeated induced sputum specimens, and findings showed that Xpert detected 76% of culture-proven TB cases, as opposed to 38% of cases detected using microscopy. The study suggests that Xpert is more sensitive than smear microscopy in detecting pulmonary TB in the children. However, almost a quarter of children with culture-confirmed TB were negative on Xpert testing, with an even higher proportion in smear-negative culture-positive children. Sixty-five percent of the children put on TB treatment in the study had both a negative culture and a negative Xpert test. Interestingly, the GeneXpert tool performed more accurate diagnosis of TB in HIV-positive children than on HIV-negative children. However, the study authors found that the number of children with HIV and culture-proven TB was too small to confirm whether sensitivity was indeed increased. Additionally, the study was inconclusive in determining enhanced rifampicin resistance as too few rifampicin-resistant cases were detected. Study authors conclude that culture should not yet be replaced with Xpert, but that it is superior to microscopy for rapid diagnosis, and critical for the detection of MDR-TB.
Another recent study in children evaluating the performance of Xpert in an HIV/TB-endemic setting in southwest Tanzania provided similar data and confirmed accuracy of Xpert in identifying smear-positive and smear-negative culture-confirmed TB cases.[71] While encouraging, these studies are the first of their kind and more evaluation of Xpert in children is needed in varied settings to conclusively prove positive findings.
Recommendations
Point-of-Care Test
Millions of TB cases go undiagnosed each year because of the ineffectiveness, inaccessibility, or expense of current diagnostic technologies. The development of a true dipstick POC test that is rapid and affordable can be used at any location where health care is provided and does not require electricity or specialist training is the ideal TB diagnostic tool that would revolutionize TB control efforts worldwide.
Researchers estimate that if widely implemented, a POC with 100% accuracy could save 625,000 lives per year, and a test with only 85% sensitivity and 97% specificity might save 392,000 lives, or 22.4% of the current annual worldwide deaths attributable to TB.[72],[73]
As such, there is an urgent need to accelerate investment to identify biomarkers that can detect those at risk for progression from TB infection to active disease, and biomarkers correlated with disease, cure, and drug resistance. Recognizing this need, several funders such as the Bill & Melinda Gates Foundation and the NIH have recently poured resources into the TB diagnosis biomarker research pipeline and a number of novel—but still nascent—technologies have since been identified. This is a start, but more can and needs to be done. Greater resource mobilization is needed to develop a low-cost and effective POC diagnostic test that can be used in all settings. Public and private sponsors need to collaborate to invest in TB diagnostic development.
Preserve and Modernize TB Sample Banks
Well-characterized specimens from people with and without TB and at various stages of disease and cure will be required to discover, develop, and validate an effective TB point-of-care test.[74] These sample banks need to be operated efficiently and have a clear open-access policy to facilitate the identification and validation of biomarkers.
In 2010, the U.S. Food and Drug Administration provided seed funds for a sample bank called the Consortium for TB Biomarkers (CTB2). CTB2 is hosted by the TB Alliance and works in partnership with the U.S. Centers for Disease Control and Prevention’s Tuberculosis Trials Consortium (TBTC) and the AIDS Clinical Trials Group of the National Institute of Allergy and Infectious Diseases (NIAID). Since its inception, CTB2 has gained considerable capacity to collect and process well-characterized samples, which are stored in an international repository and available for researchers who are investigating TB disease, treatment, and cure.
In the spring of 2012, NIAID awarded the consortium an additional five-year grant to support the discovery of new TB biomarkers.[75] Without more reliable TB biomarkers, TB clinical trial patients must be closely monitored for relapse for up to a year following treatment, which greatly increases the length and cost of TB clinical trials. The discovery of TB biomarkers could dramatically revolutionize TB clinical trial research by enabling researchers to more quickly distinguish patients who have been cured and those who are at risk of relapse. A biomarker discovery would go a long way toward enabling the testing of improved drug regimens in a faster, more effective way.
The ongoing support for CTB2 is a positive development; however, the only other sample bank in the world—the WHO’s Special Programme for Research and Training in Tropical Diseases (TDR)—is in serious jeopardy of being closed due to budget cuts and lack of prioritization. The TDR specimen bank has an essential role to play in facilitating the discovery and validation of novel biomarkers, as well as in the development of new diagnostic tests. While there remain some institutional challenges with the current setup of the specimen bank—particularly in relation to specimen collection and distribution—these problems can and ought to be fixed. The bank should be expanded rather than closed so that a wider variety of samples—such as those for pediatric TB and non-sputum samples—can becollected to support research required for the development of new diagnostics.
Enhance Uptake of the Xpert MTB/RIF Test by Reducing Machine and Cartridge Prices while Decentralizing the Test’s Availability
Despite the unprecedented global interest in and need for the Xpert MTB/RIF test, rollout of the technology has been severely hampered by the extremely high cost of the machines and cartridges. Further, maintenance of the machines when placed on the ground is expensive, complicated, and slow. As of this writing, a market intervention deal has been reached between Cepheid and UNITAID, along with the U.S. government and the Bill & Melinda Gates Foundation, to reduce pricing and expand rollout of the GeneXpert machines.
According to Cepheid, the manufacturer of the device, price reductions were nonnegotiable until a set volume of machines and cartridges were sold; however, since the machines and cartridges were too expensive to buy and maintain, the volume needed to realize the price decrease was never met. In an attempt to break this impasse, UNITAID brokered a multimillion-dollar agreement to scale up access to Xpert via a strategic market intervention. The deal made between UNITAID, the Bill & Melinda Gates Foundation, the U.S. government, and Cepheid in June 2012 included a one-time US$11.1 million buy-down payment that triggered the needed volume, thereby reducing the cost of the cartridges from US$17.00 to US$9.98 each for over 145 public-sector and NGO purchasers in low- and middle-income countries worldwide.
This collaborative market intervention is an important achievement but more still needs to be done to increase access to this revolutionary diagnostic device. Civil-society advocates have called on the manufacturer to further bring down the price of the machines and cartridges to US$7 to enable greater access to the lifesaving diagnostic. Additionally, they have proposed a tiered pricing system that would enable private-sector providers in TB-endemic settings to have access to the system. Advocates have also called for increased transparency on the current manufacturing cost of the machines and cartridges. Finally the scale-up of Xpert in South Africa—where it is being installed in every district, but administered by the NHLS, a quasi-governmental organization separate from the public health services, indicates that there is still too much lag time between test read-out and communication of test results to providers and patients, meaning that appropriate treatment may in some cases be delayed. Thus, further decentralization of Xpert and its integration directly into public health facilities is warranted.
Address Regulatory Gaps in TB Diagnostics
Accessible, inexpensive, and quality-assured TB diagnostics continue to remain elusive in high-TB-burden settings around the world. Additionally, poor regulation of the TB diagnostics market in some TB-endemic areas continues to hamper accurate diagnosis, lengthening the time to effective treatment and cure.
As referenced in last year’s pipeline report commercial serological tests for TB antibody detection are being used in at least 17 of 22 high-TB-burden countries, despite evidence of their poor performance, and though no international guideline recommends their use.[76]
Further, it is estimated that 1.5 million serological tests were done every year in India at a conservative cost estimate of US$15 million—most of which was borne by patients.[77] Further, the TDR conducted an evaluation of the performance of 19 commercially available rapid antibody detection tests for the diagnosis of TB, and found that the sensitivity of all the tests was very low, the highest being 59.7%.[78] As indicated in last year’s pipeline report, because of these data, STAG-TB passed a negative recommendation against the use of commercial serological tests for TB in 2010.[79],[80],[81]
In 2012, FIND and Becton, Dickinson and Company (BD) launched an initiative with the private Indian Kasturba Medical College (KMC) to assist in the promotion of accurate diagnosis of MDR-TB in HIV-positive people in the northern region of Karnataka.[82] The 18-month-long BD/FIND/Kasturba collaboration works by increasing access to BD’s BACTEC MGIT system at a lower price and by providing greater access to technical expertise. The collaboration will also enable KMC to become accredited to perform culture- and drug susceptibility testing. These types of innovative public-private collaborations allow access to reliable TB diagnostic equipment and promote initiation of appropriate treatment regimens from onset, thereby reducing the spread of disease.
More still needs to be done—information about WHO-recommended TB diagnostic tests and algorithms needs wider dissemination, particularly to TB providers and civil-society organizations to promote the proper use of good tests and procedures, and to prevent the use of inaccurate diagnostics.[83] Regulation of diagnostics in high-burden countries needs to be improved, and incentives are needed to encourage the private sector to replace serological tests with WHO-endorsed tools.[84]
Conclusion
The future of biomarker-driven assays and technological platforms to diagnose and guide therapeutic development and treatment of TB is very promising—particularly with molecular TB diagnosis and drug susceptibility testing. As TB pathogenesis research efforts improve, we are getting closer to finding a true point of care tool that will accurately diagnose TB. The technical and financial hurdles that need to be overcome to realize the end goal of a rapid, point-of-care test are substantial, but not insurmountable. However, a meaningful commitment to stopping the TB epidemic cannot be met without an increased, sustained research agenda for the development of new diagnostics. The future holds great promise that can only be met by rapidly improving and investing in TB diagnosis technology for the millions of people with TB worldwide.
